Deep Convolutional Networks on Graph-Structured Data
Motivation
To generalize ConvNets to high-dimensional general datasets where the graph structure is not known a priori. In this context, learning the graph structure amounts to estimating the similarity matrix, which has complexity O(N2). The authors also attempt to experimentally answer the question whether the graph estimation followed by graph convolutions offers advantages with respect to learning directly from the data with fully connected layers.
Generalizing Convolutions to Graphs
Spectral Networks
Let W be a N×N similarity matrix representing an undirected graph G, and let L=I−D−21WD−21 be its graph Laplacian where D=W⋅1. Let U=(u1,⋯,uN) be the eigenvectors of L. Then a graph convolution of input signals x∈RN with filters g on G is defined by x∗Gg=UT(Ux⊙Ug), where ⊙ represents a point-wise product. As for the theory behind the construction, see Bruna et al., 2014.
With such a construction, learning filters on a graph thus amounts to learning spectral multipliers wg=(w1,⋯,wN) with wg in
Extending the convolution to inputs x with multiple channels can be done by applying U on each channel and then using multipliers wg=(wi,j:i≤N,j≤M).
Note that one could simply replace U by an arbitrary unitary matrix, which is then optimized by BP directly. In this case the number of parameters will be O(N2).
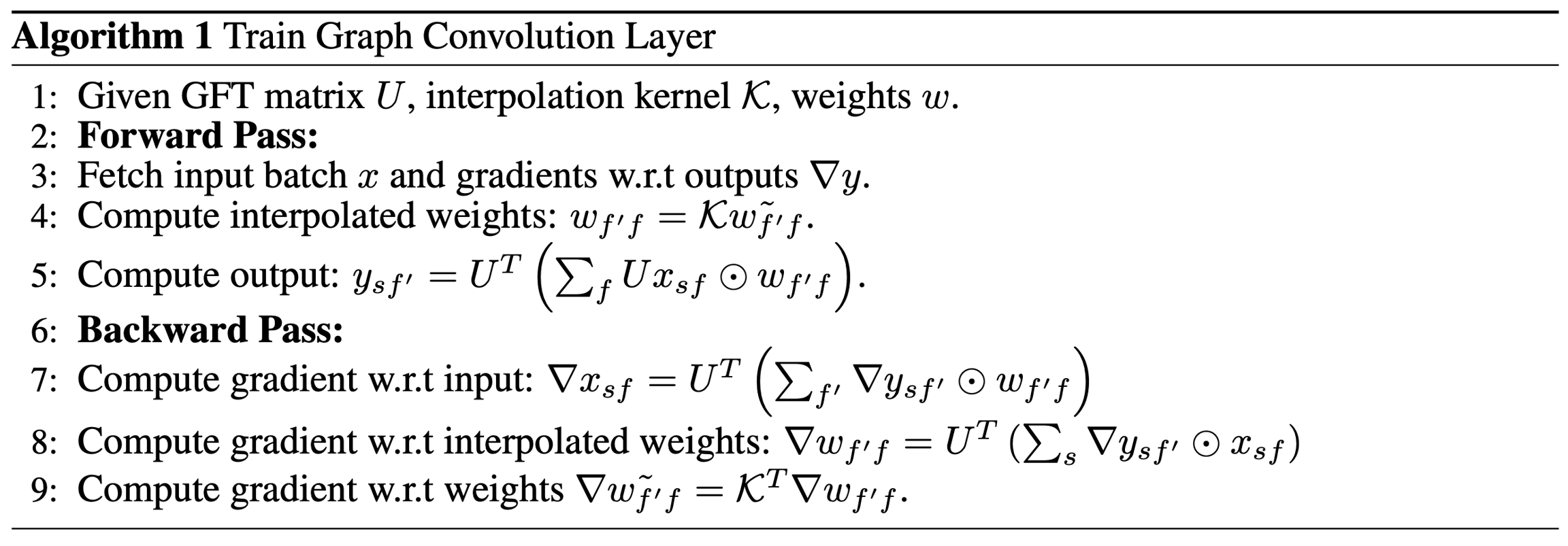
One problem with this formulation is that the multipliers will have O(N) parameters while the filter in ConvNet has O(1) parameters. To address this issue, it is necessary to restrict the class of spectral multipliers to those corresponding to localized filters. The theoretical explanation in the paper was not clear to me (Mufei), but their proposal is that we can use
K∈RN×N0 is a smoothing kernel like splines and w~g∈RN0×M.
According to Gunn 1998, spline kernel is a popular choice for interpolation modeling. A finite spline of order p, with knots located at τs is given by
where (a)+=max{a,0}.
Pooling with Hierarchical Graph Clustering
The authors construct neighborhoods at different scales using multi-resolution spectral clustering and consider both average and max-pooling as in standard ConvNets.
Graph Construction
When our non-Euclidean dataset does not have a prior graph structure, we need to estimate the similarity matrix W before constructing the spectral network. The authors presented two approaches for graph construction, one unsupervised by measuring joint feature statistics, and another one supervised using an initial network as a proxy for the estimation.
Unsupervised Graph Estimation
Given X∈RN×M, N is the number of samples and M is the number of features, a distance can be defined between a pair of features i, j by
where Xi is the i-th column of X. This definition can be problematic as it fails to capture higher-order statistics and might be fragile when the features are not normalized. To address these issues, we can replace the Euclidean distance by the Z-score (thus renormalizing each feature by its standard deviation), the "square-correlation" (computing the correlation of squares of previously whitened features), or the mutual information.
A Gaussian diffusion kernel can be built from the chosen distance:
In experiments, the authors consider the variant of self-tuning diffusion kernel
where σi is computed as the distance d(i,ik) corresponding to the k-th nearest neighbor ik of feature i. This defines a kernel whose variance is locally adapted around each feature point, as opposed to the previous one where the variance is shared.
Supervised Graph Estimation
Given a training set with normalized features (the authors used feature normalization by its standard deviation and they suggested the possibility of whitening completely the data) X∈RN×M and labels y∈{1,⋯,C}L, they initialized a FNN ϕ with K layers of weights W1,⋯,WK with ReLU and dropout. They then extracted the first layer features W1∈RN×M1, where M1 is the number of first-layer hidden features and consider the distance
The rest can be performed as in the unsupervised construction.
Last updated