Layerwise Adaptive Sampling
tags: GCN, sampling, NeuralPS2018
Adaptive Sampling Towards Fast Graph Representation Learning
Multiple vertices may have some common neighbors so neighbor sampling can result in repeated samples. We can avoid the over-expansion and accelerate the training of GCN by controlling the size of the sampled neighborhoods in each layer.
The core of the method is to define an appropriate sampler for the layer-wise sampling. A common objective to design the sampler is to minimize the resulting variance. Unfortunately, the optimal sampler to minimize the variance is uncomputable due to the inconsistency between the top-down sampling and the bottom-up propagation in our network. To tackle this issue a parametrized sampler is used and the resulting variance, being a loss term, can be directly optimized with BP.
The authors also propose to include skip connection for message passing so that 2nd order proximity
To sum up, the contributions include:
Overexpansion of neighborhood ⇒ Layer-wise neighbor sampling
Uncomputable optimal sampler ⇒ Feature based parametrized sampler + directly optimizing variance in the objective
Preserving 2nd order proximity ⇒ Skip connection for message passing
Related Work
Spectral: defines the convolution operation in Fourier domain
Spectral networks and locally connected networks on graphs: defines the convolution operation in Fourier domain
Deep convolutional networks on graph-structured data: enables localized filtering by applying efficient spectral filters
Convolutional neural networks on graphs with fast localized spectral filtering: employs Chebyshev expansion of the graph Laplacian to avoid the eigendecomposition
GCN: simplify previous methods with first-order expansion and re-parameterization trick
Non-spectral: define convolution on graph by using the spatial connections directly
Convolutional neural networks on graphs for learning molecular fingerprints: learns a weight matrix for each node degree
Diffusion-convolutional neural networks: defines multiple-hop neighborhoods by using the power series of a transition matrix
Learning convolutional neural networks for graphs: extracts normalized neighborhoods that contain a fixed number of nodes
Leap of model capacity: implicitly weight node importance of a neighborhood
Geometric deep learning on graphs and manifolds using mixture model cnns: build ConvNet on graphs using the patch operation
GAT: compute the hidden representations of each node on graph by attending over its neighbors following a self-attention strategy
Method Formulation
Monte Carlo GCN
In GCN, the update rule for node features is
where
We can reformulate the rule above with expectation:
where
The term Eu∼p(u∣v)[hu(l)] may be approximated with Monte Carlo sampling:
with u1,⋯,un sampled with p(u∣v).
Layer-wise sampling
The layer-wise sampling can be incorporated with importance sampling. Assume q(u∣v1,⋯,vm) is a conditional layer-wise sampling distribution, which we will introduce later. The original update rule is equivalent to
where v∈{v1,⋯,vm}. We can also perform Monte Carlo sampling as above.
Opposed to the node-wise Monte Carlo method where the nodes are sampled independently for each vi, the layer-wise sampling is performed only once for v1,⋯,vm. As a result, the total number of sampling nodes only grows linearly with the network depth if we fix the sampling size n.
Variance reduction
For simplicity we abbreviate q(u∣v1,⋯,vm) as q(u). For a good choice of q(u), we seek to reduce the induced variance of μ^q(vj)=n1∑i=1nq(ui)p(ui∣vj)hui(l), vj∈{v1,⋯,vm}, ui's are sampled from q(u).
Note that there is a small problem with the original argument of the authors where they treat hui(l) as a scalar rather than a vector. The argumentation there was more for some motivation.
If hui(l) is a scalar, then as mentioned in page 6, Chapter 9 Importance Sampling, Monte Carlo theory, methods and examples, the variance is
and the optimal q in terms of variance is given by
For both reasons that: 1. hui(l) is a vector and we want a scalar 2. Even the above holds, Ep[∣hui(l)∣] cannot be evaluated efficiently.
The authors use a linear layer g(x(ui))=Wgx(ui) to replace hui(l) in the eqution above, where Wg∈R1×D. x(ui) is the node feature. We can then perform a Monte Carlo estimation
To make the estimation independent of vj for the purpose of layerwise sampling, we can do
To make the variance reduction process adaptive, we can directly add the estimated variance to the objective function:
where u1,⋯,un are sampled from q.
Skip Connections
For the nodes of the (l+1) layer, we can add direct connections between them and the nodes in the (l−1) layer for feature update.
where:
{sj}j=1n are nodes sampled in the (l−1)-th layer.
a^skip(vi,sj) is approximated by ∑k=1na^(vi,uk)a^(uk,sj) where u1,⋯,uk are nodes sampled in the l-th layer.
Wskip(l−1)=W(l−1)W(l), where W(l) and W(l−1) are the filters of the l-th and (l−1)-th layers
The final update is then
Attention
In GAT, A^(u,v) is replaced by SoftMax(LeakyReLU(W1h(l)(vi),W2h(l)(uj))), the dependence on h(l)(vi),h(l)(uj) makes it intractable for the method proposed as the sampling of nodes in the l th layer depends on the nodes in the l+1 th layer. The author proposed to use
instead, where W1 and W2 and x(vi),x(uj) are the node features.
Experiments
Categorizing academic papers in the citation network datasets -- Cora (O(103) nodes), Citeseer (O(103) nodes) and Pubmed (O(104) nodes)
Predicting which community different posts belong to in Reddit (O(105) nodes)
The sampling framework is inductive (separates out test data from training) rather than transductive (all vertices are included). In test time, the authors do not use sampling.

The experiments are conducted with random seeds over 20 trials with mean and standard variances recorded.
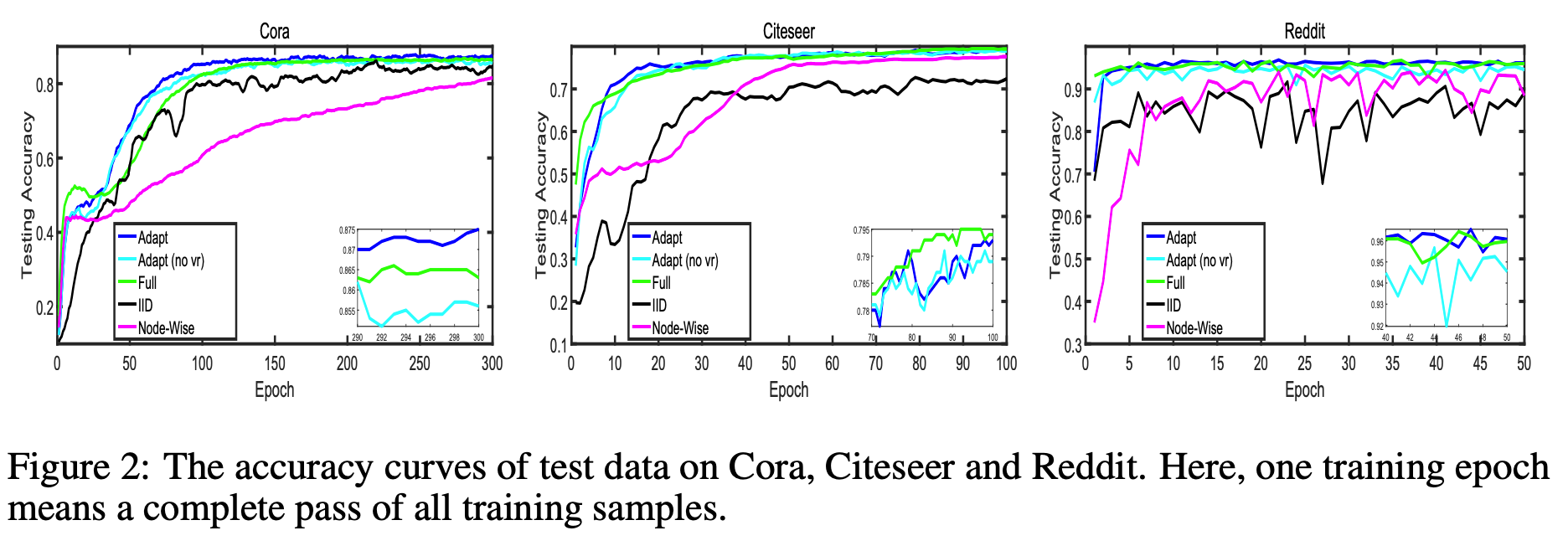
From the learning curves, the performance using layer-wise sampling is comparable to the full training and is better than Node-Wise(re-implementation of GraphSage) and IID(re-implementation of FastGCN). However, the variance reudction seems not to help much here.
Note the leanrning curves above are based on a re-implementation of FastGCN and GraphSage for a fair comparison. The comparison with the official implementation is presented below:


For skip connections, the authors also add a skip connection between the first layer and the last layer. I think skip connection does not help too much on the datasets concerned.

Last updated