GAT
Summary
Incorporate multi-head attention into GCN, which clearly strengthens its model capacity. The authors claim that this is helpful for inductive learning: where we want to generalize a graph neural network to unseen nodes and graphs.
Attention
The key difference between GAT and GCN is how to aggregate the information from the one-hop neighbourhood.
For GCN, a convolution operation produces the normalized sum of the node features of neighbours.
where N(i) is the union of vi and all its one-hop neighbours, cij=∣N(i)∣1∣N(j)∣1 is a normalization constant based on graph structure, σ is an activation function such as ReLU, W(l) is a shared weight matrix for node-wise feature transformation.
GAT introduces the attention mechanism as substitute to the statically normalized convolution operation to remove its dependency on graph structure. It aggregates the neighborhood features by assigning different weights to features from neighbor nodes depending on the features of the current node.

where ∥ is the concatenation operation, σ(⋅) is some activation function, hi(l)∈RF, W∈RF′×F, aT∈R1×2F′. Both W and a are learnable.
Note that employing attention mechanism allows dealing with directed graphs.
Multi-head Attention
Analogous to multiple channels in ConvNet, GAT introduces multi-head attention to enrich the model capacity. The authors claimed that this is beneficial for stabilizing the learning process of self-attention. Basically we can have multiple "suits" of parameters for computing αij and W(l) in hi(l+1)=σ(∑j∈N(i)αijW(l)hj(l)).
Formally this can be done by first performing computation for each head and then aggregate the result from each head with
or
where K is the number of heads. The authors suggest using concatenation for intermediary layers and average for the final layer.
Experiments
Datasets
Transductive Learning: They used 3 standard citation benchmark datasets: Cora, Citeseer and Pubmed. The node features correspond to elements of a bag-of-words representation of a document. Each node has a class label. They allow 20 nodes per class to be used for training but the training algorithm has access to all of the nodes' feature vectors. The test set consists of 1000 test nodes and the validation set consists of 500 nodes.
Inductive Learning: They make use of a protein-protein interaction (PPI) dataset that consists of graphs corresponding to different human tissues. For inductive learning, testing graphs remain completely unobserved during training. The node features are composed of positional gene sets, motif gene sets and immunological signatures. There are 121 labels for each node set from gene ontology, and a node can possess several labels simultaneously.
Below is a summary of dataset statistics:

State-of-the-art Methods
Transductive Learning:
label propagation
semi-supervised embedding
manifold regularization
skip-gram based graph embeddings
iterative classification algorithm
planetoid
GCN
ChebyNet
MoNet
MLP where no graph structure incorporated
Inductive Learning:
GraphSAGE-GCN (extends graph convolution to inductive learning): hi(l+1)=σ(∣N(i)∣1∑j∈N(i)W(l)hj(l))
GraphSAGE-mean (mean pooling)
GraphSAGE-LSTM
GraphSAGE-pool (max pooling)
MLP where no graph structure incorporated
Experimental Setup
Transductive Learning:
Model: two-layer GAT
Hyperparameters: optimized based on Cora
Model flow:
8 head attention with F′=8 features for each head
An exponential linear unit (ELU)
Classification layer: linear layer with C out features corresponding to C classes, followed by a softmax activation
Regularization: L2 with λ=0.0005
Dropout: p=0.6 applied to both layers' inputs as well as to the normalized attention coefficients (at each training iteration, each node is exposed to a stochastically sampled neighborhood).
For Pubmed, 8 output attention heads are applied (rather than 1), with λ=0.001 for regularization
Inductive Learning:
Model: three-layer GAT
Model flow:
For the first two layers, 4 attention heads are used with F′=256.
An exponential linear unit (ELU)
(multi-label) classification layer: 6 attention heads computing 121 features each, that are averaged and followed by a logistic sigmoid activation
Dataset is large enough so no dropout or regularization is required
Skip connections across the intermediate attention layer
Batch size 2 during training
Both models use:
Glorot (Xavier) initialization
cross-entropy objective
Adam with initial learning rate of 0.01 for Pubmed and 0.005 for all other datasets
Early stopping on both the cross-entropy loss and accuracy (transductive) or micro-F1 (inductive) score on the validation nodes, with a patience of 100 epochs
For the transductive tasks, they report the mean accuracy (with standard deviation) on the test nodes after 100 runs. For the Chebyshev filter-based approach, they provide the maximum reported performance for filters of orders K=2 and K=3. For a fair comparison, they further evaluate a GCN model that computes 64 hidden features, attempting both the ReLU and ELU activation, and reporting the better result after 100 runs.
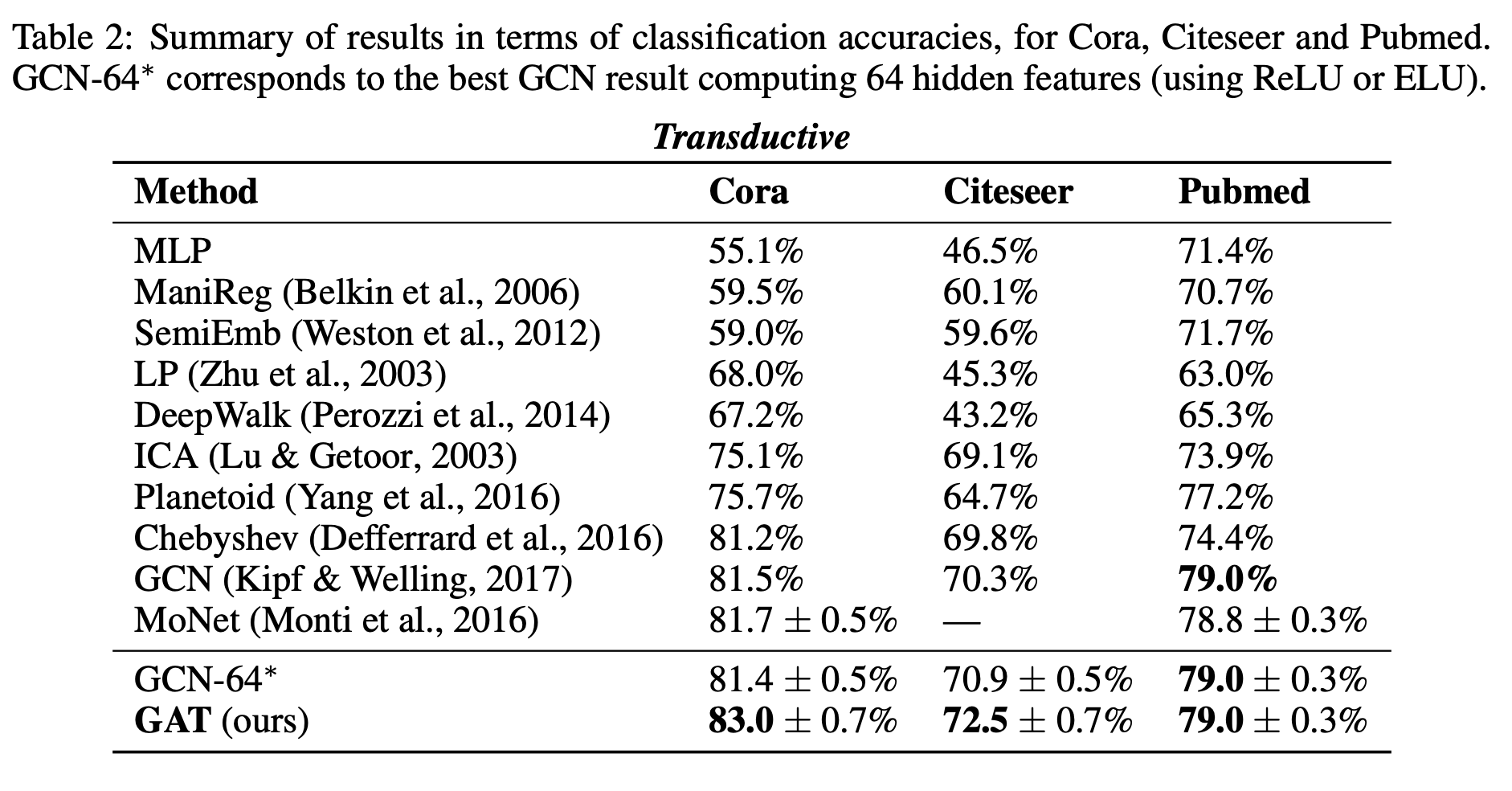
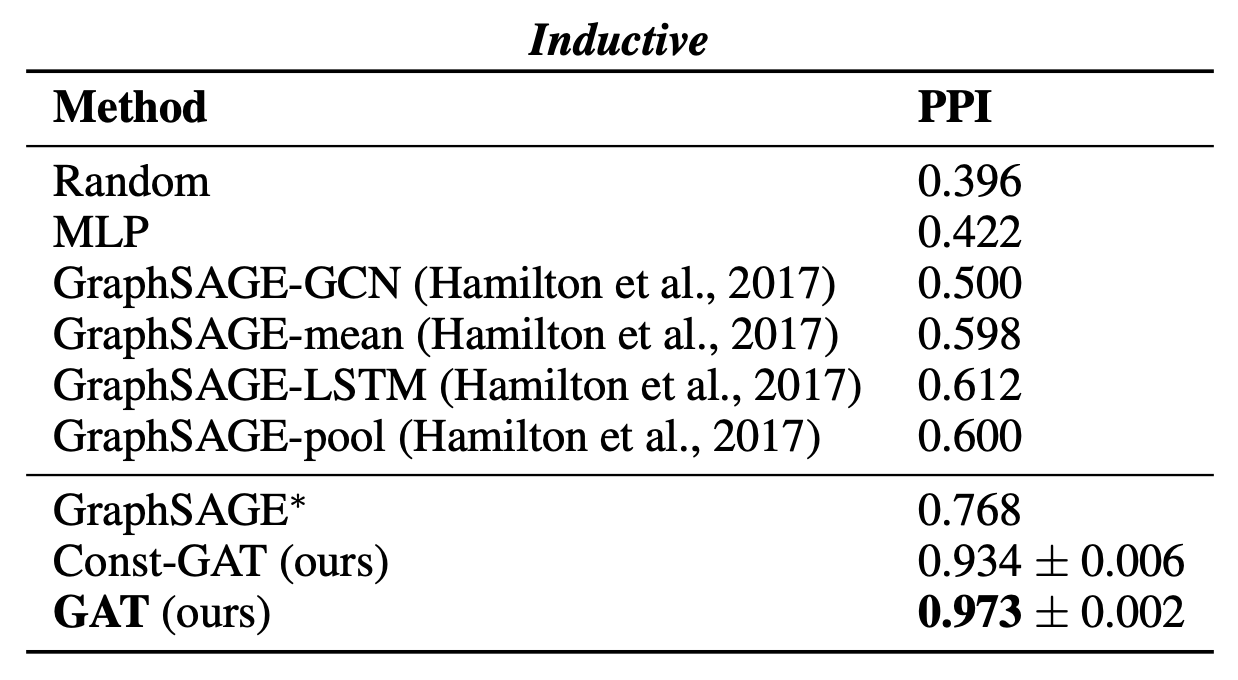
For the inductive task, they reported the micro-averaged F1 score on the nodes of the two unseen test graphs, averaged after 10 runs. When comparing against GraphSAGE, they retune the hyperparameters and denote that variant by GraphSAGE∗ They also tried a GAT variant with constant attention (1 for each node), denoted by Const-GAT.
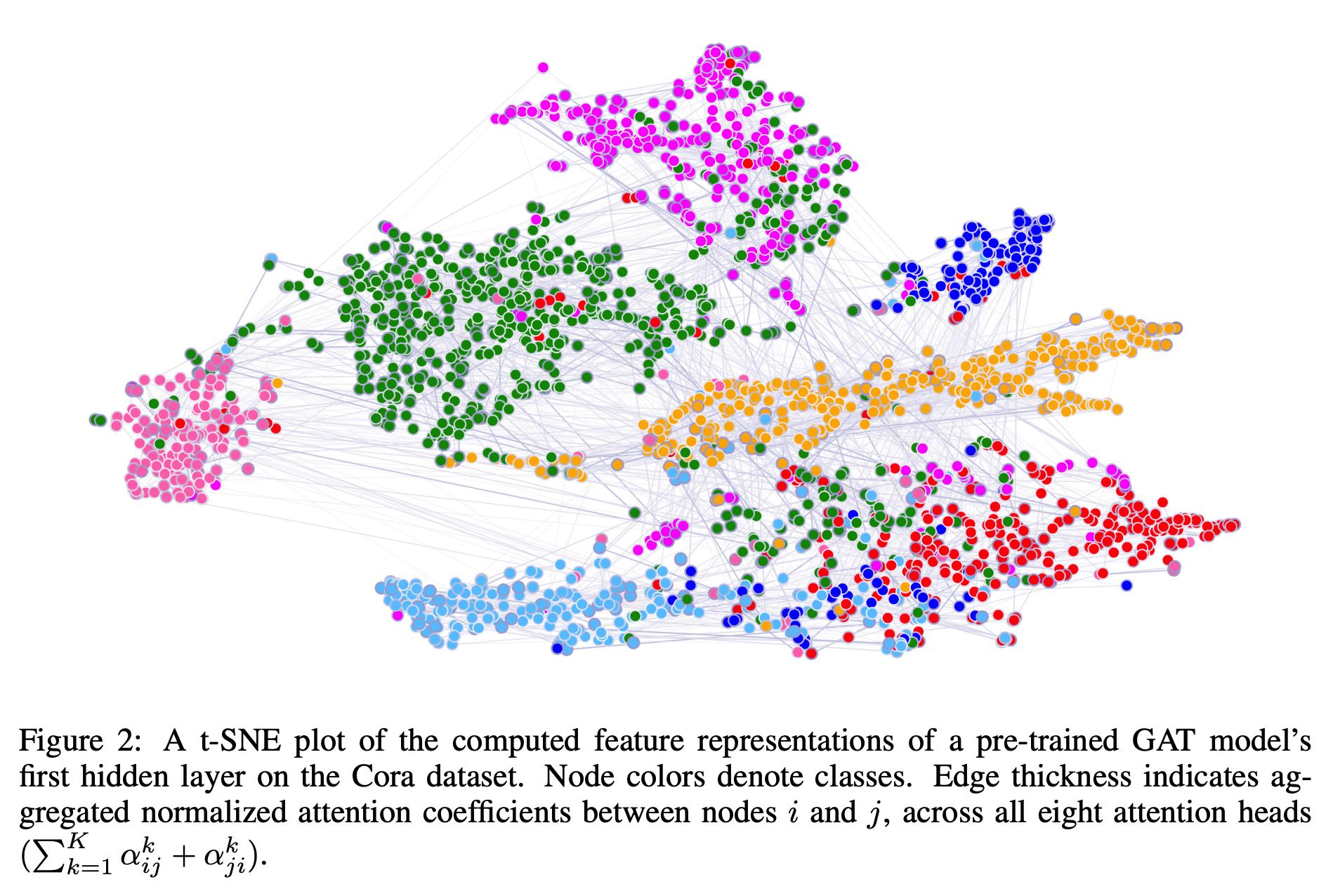
The authors also visualized the representations extracted by the first layer of a GAT pre-trained on the Cora with t-SNE.
Last updated